| Citation: | Pirone D, Sirico D, Miccio L, Bianco V, Mugnano M et al. 3D imaging lipidometry in single cell by in-flow holographic tomography. Opto-Electron Adv 6, 220048 (2023). doi: 10.29026/oea.2023.220048 |
3D imaging lipidometry in single cell by in-flow holographic tomography
-
Abstract
The most recent discoveries in the biochemical field are highlighting the increasingly important role of lipid droplets (LDs) in several regulatory mechanisms in living cells. LDs are dynamic organelles and therefore their complete characterization in terms of number, size, spatial positioning and relative distribution in the cell volume can shed light on the roles played by LDs. Until now, fluorescence microscopy and transmission electron microscopy are assessed as the gold standard methods for identifying LDs due to their high sensitivity and specificity. However, such methods generally only provide 2D assays and partial measurements. Furthermore, both can be destructive and with low productivity, thus limiting analysis of large cell numbers in a sample. Here we demonstrate for the first time the capability of 3D visualization and the full LD characterization in high-throughput with a tomographic phase-contrast flow-cytometer, by using ovarian cancer cells and monocyte cell lines as models. A strategy for retrieving significant parameters on spatial correlations and LD 3D positioning inside each cell volume is reported. The information gathered by this new method could allow more in depth understanding and lead to new discoveries on how LDs are correlated to cellular functions.-
Keywords:
- lipid droplets /
- label-free phase-contrast imaging /
- in-flow tomography /
- 3D imaging
-
-
References
[1] Murphy DJ. The dynamic roles of intracellular lipid droplets: from archaea to mammals. Protoplasma 249, 541–585 (2012). doi: 10.1007/s00709-011-0329-7 [2] Olzmann JA, Carvalho P. Dynamics and functions of lipid droplets. Nat Rev Mol Cell Biol 20, 137–155 (2019). doi: 10.1038/s41580-018-0085-z [3] Welte MA, Gould AP. Lipid droplet functions beyond energy storage. Biochim Biophys Acta - Mol Cell Biol Lip 1862, 1260–1272 (2017). doi: 10.1016/j.bbalip.2017.07.006 [4] Wu HX, Carvalho P, Voeltz GK. Here, there, and everywhere: the importance of ER membrane contact sites. Science 361, eaan5835 (2018). doi: 10.1126/science.aan5835 [5] Melo RCN, Weller PF. Lipid droplets in leukocytes: organelles linked to inflammatory responses. Exp Cell Res 340, 193–197 (2016). doi: 10.1016/j.yexcr.2015.10.028 [6] Geltinger F, Schartel L, Wiederstein M, Tevini J, Aigner E et al. Friend or foe: lipid droplets as organelles for protein and lipid storage in cellular stress response, aging and disease. Molecules 25, 5053 (2020). doi: 10.3390/molecules25215053 [7] Imai Y, Cousins RS, Liu SM, Phelps BM, Promes JA. Connecting pancreatic islet lipid metabolism with insulin secretion and the development of type 2 diabetes. Ann N Y Acad Sci 1461, 53–72 (2020). doi: 10.1111/nyas.14037 [8] Williams KJ, Tabas I. The response-to-retention hypothesis of early atherogenesis. Arterioscler, Thromb, Vasc Biol 15, 551–561 (1995). doi: 10.1161/01.ATV.15.5.551 [9] Gluchowski NL, Becuwe M, Walther TC, Farese RV Jr. Lipid droplets and liver disease: from basic biology to clinical implications. Nat Rev Gastroenterol Hepatol 14, 343–355 (2017). doi: 10.1038/nrgastro.2017.32 [10] Bosch M, Sánchez-Álvarez M, Fajardo A, Kapetanovic R, Steiner B et al. Mammalian lipid droplets are innate immune hubs integrating cell metabolism and host defense. Science 370, eaay8085 (2020). doi: 10.1126/science.aay8085 [11] Liu L, Zhang K, Sandoval H, Yamamoto S, Jaiswal M et al. Glial lipid droplets and ROS induced by mitochondrial defects promote neurodegeneration. Cell 160, 177–190 (2015). doi: 10.1016/j.cell.2014.12.019 [12] Santos CR, Schulze A. Lipid metabolism in cancer. FEBS J 279, 2610–2623 (2012). doi: 10.1111/j.1742-4658.2012.08644.x [13] Cruz ALS, de A. Barreto E, Fazolini NPB, Viola JPB, Bozza PT. Lipid droplets: platforms with multiple functions in cancer hallmarks. Cell Death Dis 11, 105 (2020). doi: 10.1038/s41419-020-2297-3 [14] Tirinato L, Pagliari F, Limongi T, Marini M, Falqui A et al. An overview of lipid droplets in cancer and cancer stem cells. Stem Cells Int 2017, 1656053 (2017). doi: 10.1155/2017/1656053 [15] Thun MJ, Jacobs EJ, Patrono C. The role of aspirin in cancer prevention. Nat Rev Clin Oncol 9, 259–267 (2012). doi: 10.1038/nrclinonc.2011.199 [16] Bozza PT, Melo RCN, Bandeira-Melo C. Leukocyte lipid bodies regulation and function: contribution to allergy and host defense. Pharmacol Ther 113, 30–49 (2007). doi: 10.1016/j.pharmthera.2006.06.006 [17] den Brok MH, Raaijmakers TK, Collado-Camps E, Adema GJ. Lipid droplets as immune modulators in myeloid cells. Trends Immunol 39, 380–392 (2018). doi: 10.1016/j.it.2018.01.012 [18] Monson EA, Trenerry AM, Laws JL, Mackenzie JM, Helbig KJ. Lipid droplets and lipid mediators in viral infection and immunity. FEMS Microbiol Rev 45, fuaa066 (2021). doi: 10.1093/femsre/fuaa066 [19] Dias SSG, Soares VC, Ferreira AC, Sacramento CQ, Fintelman-Rodrigues N et al. Lipid droplets fuel SARS-CoV-2 replication and production of inflammatory mediators. PLoS Pathog 16, e1009127 (2020). doi: 10.1371/journal.ppat.1009127 [20] Nardacci R, Colavita F, Castilletti C, Lapa D, Matusali G et al. Evidences for lipid involvement in SARS-CoV-2 cytopathogenesis. Cell Death Dis 12, 263 (2021). doi: 10.1038/s41419-021-03527-9 [21] Chen XX, Wu TL, Gong ZY, Guo JH, Liu XS et al. Lipid droplets as endogenous intracellular microlenses. Light Sci Appl 10, 242 (2021). doi: 10.1038/s41377-021-00687-3 [22] Martins AS, Martins IC, Santos NC. Methods for lipid droplet biophysical characterization in Flaviviridae infections. Front Microbiol 9, 1951 (2018). doi: 10.3389/fmicb.2018.01951 [23] Fam TK, Klymchenko AS, Collot M. Recent advances in fluorescent probes for lipid droplets. Materials 11, 1768 (2018). doi: 10.3390/ma11091768 [24] Lehmussola A, Ruusuvuori P, Selinummi J, Huttunen H, Yli-Harja O. Computational framework for simulating fluorescence microscope images with cell populations. IEEE Trans Med Imaging 26, 1010–1016 (2007). doi: 10.1109/TMI.2007.896925 [25] Fujimoto T, Ohsaki Y, Suzuki M, Cheng JL. Imaging lipid droplets by electron microscopy. Methods Cell Biol 116, 227–251 (2013). doi: 10.1016/B978-0-12-408051-5.00012-7 [26] Locatelli A, Iommarini L, Graziadio A, Leoni A, Porcelli AM et al. Dansyl acetyl trehalose: a novel tool to investigate the cellular fate of trehalose. RSC Adv 9, 15350–15356 (2019). doi: 10.1039/C9RA01800J [27] Wilson MH, Ekker SC, Farber SA. Imaging cytoplasmic lipid droplets in vivo with fluorescent perilipin 2 and perilipin 3 knock-in zebrafish. eLife 10, e66393 (2021). doi: 10.7554/eLife.66393 [28] Ferraro P, Alferi D, de Nicola S, de Petrocellis L, Finizio A et al. Quantitative phase-contrast microscopy by a lateral shear approach to digital holographic image reconstruction. Opt Lett 31, 1405–1407 (2006). doi: 10.1364/OL.31.001405 [29] Sandoz PA, Tremblay C, van der Goot FG, Frechin M. Image-based analysis of living mammalian cells using label-free 3D refractive index maps reveals new organelle dynamics and dry mass flux. PLoS Biol 17, e3000553 (2019). doi: 10.1371/journal.pbio.3000553 [30] Hsieh CM, Liu PY, Chin LK, Zhang JB, Wang K et al. Regulation of lipid droplets in live preadipocytes using optical diffraction tomography and Raman spectroscopy. Opt Express 27, 22994–23008 (2019). doi: 10.1364/OE.27.022994 [31] Huang YQ, Xia WJ, Lu ZX, Liu Y, Chen H et al. Noise-powered disentangled representation for unsupervised speckle reduction of optical coherence tomography images. IEEE Trans Med Imaging 40, 2600–2614 (2021). doi: 10.1109/TMI.2020.3045207 [32] Jung JH, Hong SJ, Kim HB, Kim G, Lee M et al. Label-free non-invasive quantitative measurement of lipid contents in individual microalgal cells using refractive index tomography. Sci Rep 8, 6524 (2018). doi: 10.1038/s41598-018-24393-0 [33] Kim K, Lee S, Yoon J, Heo J, Choi C et al. Three-dimensional label-free imaging and quantification of lipid droplets in live hepatocytes. Sci Rep 6, 36815 (2016). doi: 10.1038/srep36815 [34] Park S, Ahn JW, Jo Y, Kang HY, Kim HJ et al. Label-free tomographic imaging of lipid droplets in foam cells for machine-learning-assisted therapeutic evaluation of targeted Nanodrugs. ACS Nano 14, 1856–1865 (2020). doi: 10.1021/acsnano.9b07993 [35] Hong YR, Dao KP, Kim T, Lee S, Shin Y et al. Label‐free quantitative analysis of Coacervates via 3D phase imaging. Adv Opt Mater 9, 2100697 (2021). doi: 10.1002/adom.202100697 [36] Moon I, Javidi B. 3-D visualization and identification of biological microorganisms using partially temporal incoherent light in-line computational holographic imaging. IEEE Trans Med Imaging 27, 1782–1790 (2008). doi: 10.1109/TMI.2008.927339 [37] Merola F, Memmolo P, Miccio L, Savoia R, Mugnano M et al. Tomographic flow cytometry by digital holography. Light Sci Appl 6, e16241 (2017). doi: 10.1038/lsa.2016.241 [38] Villone MM, Memmolo P, Merola F, Mugnano M, Miccio L et al. Full-angle tomographic phase microscopy of flowing quasi-spherical cells. Lab Chip 18, 126–131 (2018). doi: 10.1039/C7LC00943G [39] Pirone D, Memmolo P, Merola F, Miccio L, Mugnano M et al. Rolling angle recovery of flowing cells in holographic tomography exploiting the phase similarity. Appl Opt 60, A277–A284 (2021). doi: 10.1364/AO.404376 [40] Ryu D, Ryu D, Baek Y, Cho H, Kim G et al. DeepRegularizer: rapid resolution enhancement of tomographic imaging using deep learning. IEEE Trans Med Imaging 40, 1508–1518 (2021). doi: 10.1109/TMI.2021.3058373 [41] Pirone D, Mugnano M, Memmolo P, Merola F, Lama GC et al. Three-dimensional quantitative intracellular visualization of Graphene oxide nanoparticles by tomographic flow cytometry. Nano Lett 21, 5958–5966 (2021). doi: 10.1021/acs.nanolett.1c00868 [42] Campos V, Rappaz B, Kuttler F, Turcatti G, Naveiras O. High-throughput, nonperturbing quantification of lipid droplets with digital holographic microscopy. J Lipid Res 59, 1301–1310 (2018). doi: 10.1194/jlr.D085217 [43] Yanina IY, Lazareva EN, Tuchin VV. Refractive index of adipose tissue and lipid droplet measured in wide spectral and temperature ranges. Appl Opt 57, 4839–4848 (2018). doi: 10.1364/AO.57.004839 [44] Sheneman L, Stephanopoulos G, Vasdekis AE. Deep learning classification of lipid droplets in quantitative phase images. PLoS One 16, e0249196 (2021). doi: 10.1371/journal.pone.0249196 [45] Park YK, Depeursinge C, Popescu G. Quantitative phase imaging in biomedicine. Nat Photonics 12, 578–589 (2018). doi: 10.1038/s41566-018-0253-x [46] Eden E, Waisman D, Rudzsky M, Bitterman H, Brod V et al. An automated method for analysis of flow characteristics of circulating particles from in vivo video microscopy. IEEE Trans Med Imaging 24, 1011–1024 (2005). doi: 10.1109/TMI.2005.851759 [47] Moore MJ, Sebastian JA, Kolios MC. Determination of cell nucleus-to-cytoplasmic ratio using imaging flow cytometry and a combined ultrasound and photoacoustic technique: a comparison study. J Biomed Opt 24, 106502 (2019). doi: 10.1117/1.jbo.24.10.106502 [48] Walther TC, Farese RV Jr. Lipid droplets and cellular lipid metabolism. Annu Rev Biochem 81, 687–714 (2012). doi: 10.1146/annurev-biochem-061009-102430 [49] Farese RV, Walther TC. Lipid droplets go nuclear. J Cell Biol 212, 7–8 (2016). doi: 10.1083/jcb.201512056 [50] Ohsaki Y, Sołtysik K, Fujimoto T. The lipid droplet and the endoplasmic reticulum. In Organelle Contact Sites 111–120 (Springer, 2017);http://doi.org/10.1007/978-981-10-4567-7_8. [51] Tauchi-Sato K, Ozeki S, Houjou T, Taguchi R, Fujimoto T. The surface of lipid droplets is a phospholipid monolayer with a unique fatty acid composition. J Biol Chem 277, 44507–44512 (2002). doi: 10.1074/jbc.M207712200 [52] Bersuker K, Peterson CWH, To M, Sahl SJ, Savikhin V et al. A proximity labeling strategy provides insights into the composition and dynamics of lipid droplet proteomes. Dev Cell 44, 97–112.e7 (2018). doi: 10.1016/j.devcel.2017.11.020 [53] Dannhauser D, Rossi D, Memmolo P, Finizio A, Ferraro P et al. Biophysical investigation of living monocytes in flow by collaborative coherent imaging techniques. Biomed Opt Express 9, 5194–5204 (2018). doi: 10.1364/BOE.9.005194 [54] Cotte Y, Toy F, Jourdain P, Pavillon N, Boss D et al. Marker-free phase nanoscopy. Nat Photonics 7, 113–117 (2013). doi: 10.1038/nphoton.2012.329 [55] Miccio L, Cimmino F, Kurelac I, Villone MM, Bianco V et al. Perspectives on liquid biopsy for label‐free detection of “circulating tumor cells” through intelligent lab‐on‐chips. View 1, 20200034 (2020). doi: 10.1002/VIW.20200034 [56] Schneider CA, Rasband WS, Eliceiri KW. NIH image to ImageJ: 25 years of image analysis. Nat Methods 9, 671–675 (2012). doi: 10.1038/nmeth.2089 [57] Kim MK. Principles and techniques of digital holographic microscopy. SPIE Rev 1, 018005 (2010). doi: 10.1117/6.0000006 [58] Memmolo P, Miccio L, Paturzo M, di Caprio G, Coppola G et al. Recent advances in holographic 3D particle tracking. Adv Opt Photonics 7, 713–755 (2015). doi: 10.1364/AOP.7.000713 [59] Zhou WJ, Yu YJ, Asundi A. Study on aberration suppressing methods in digital micro-holography. Opt Lasers Eng 47, 264–270 (2009). doi: 10.1016/j.optlaseng.2008.04.026 [60] Kemao Q. Windowed Fourier transform for fringe pattern analysis. Appl Opt 43, 2695–2702 (2004). doi: 10.1364/AO.43.002695 [61] Bioucas-Dias JM, Valadao G. Phase unwrapping via graph cuts. IEEE Trans Image Process 16, 698–709 (2007). doi: 10.1109/TIP.2006.888351 [62] Kak AC, Slaney M. Principles of Computerized Tomographic Imaging (Society for Industrial and Applied Mathematics, Philadelphia, 2001); http://doi.org/10.1137/1.9780898719277. -
Supplementary Information
Movie 1 
Movie 2 
-
Access History
Article Metrics
-
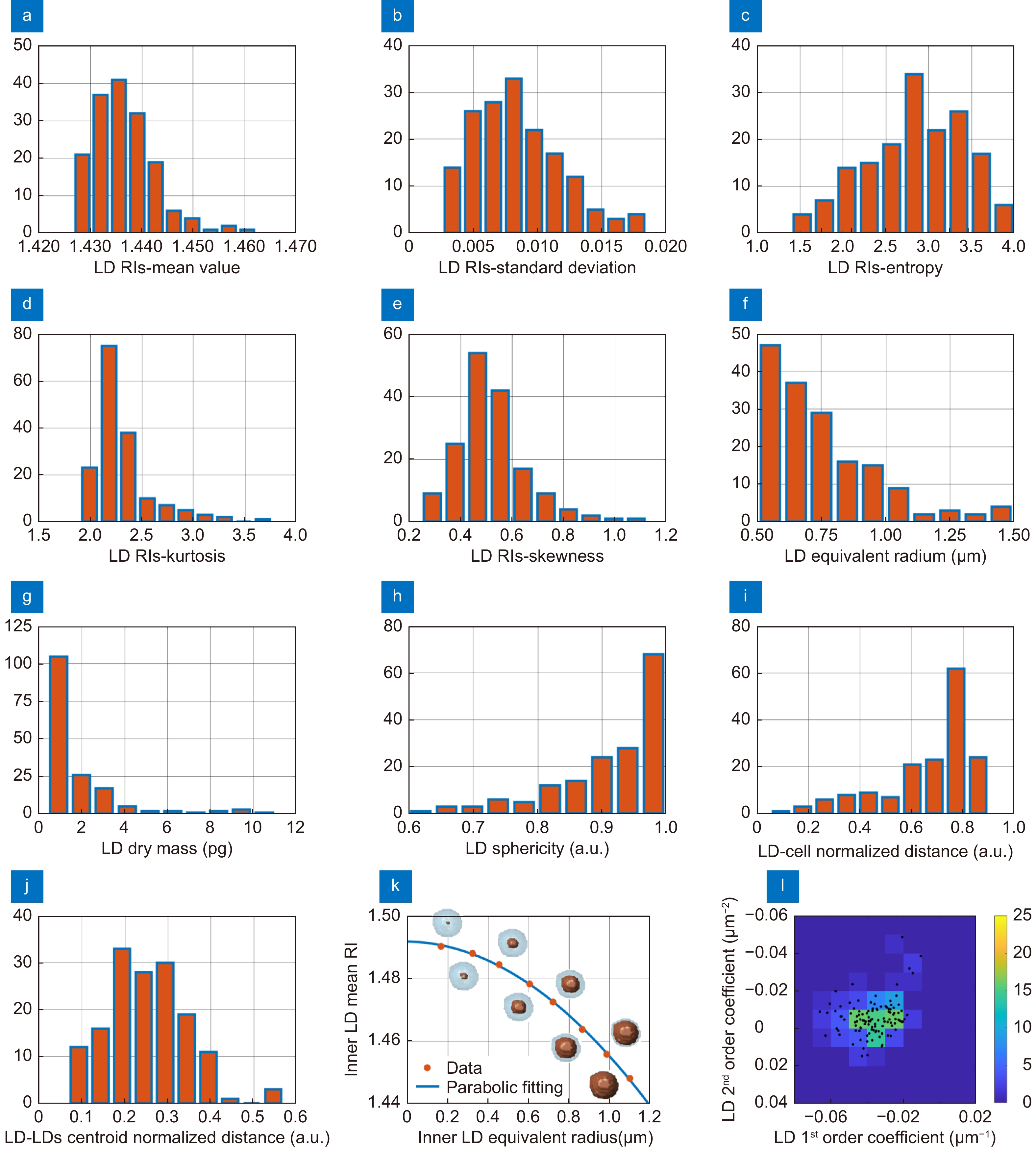
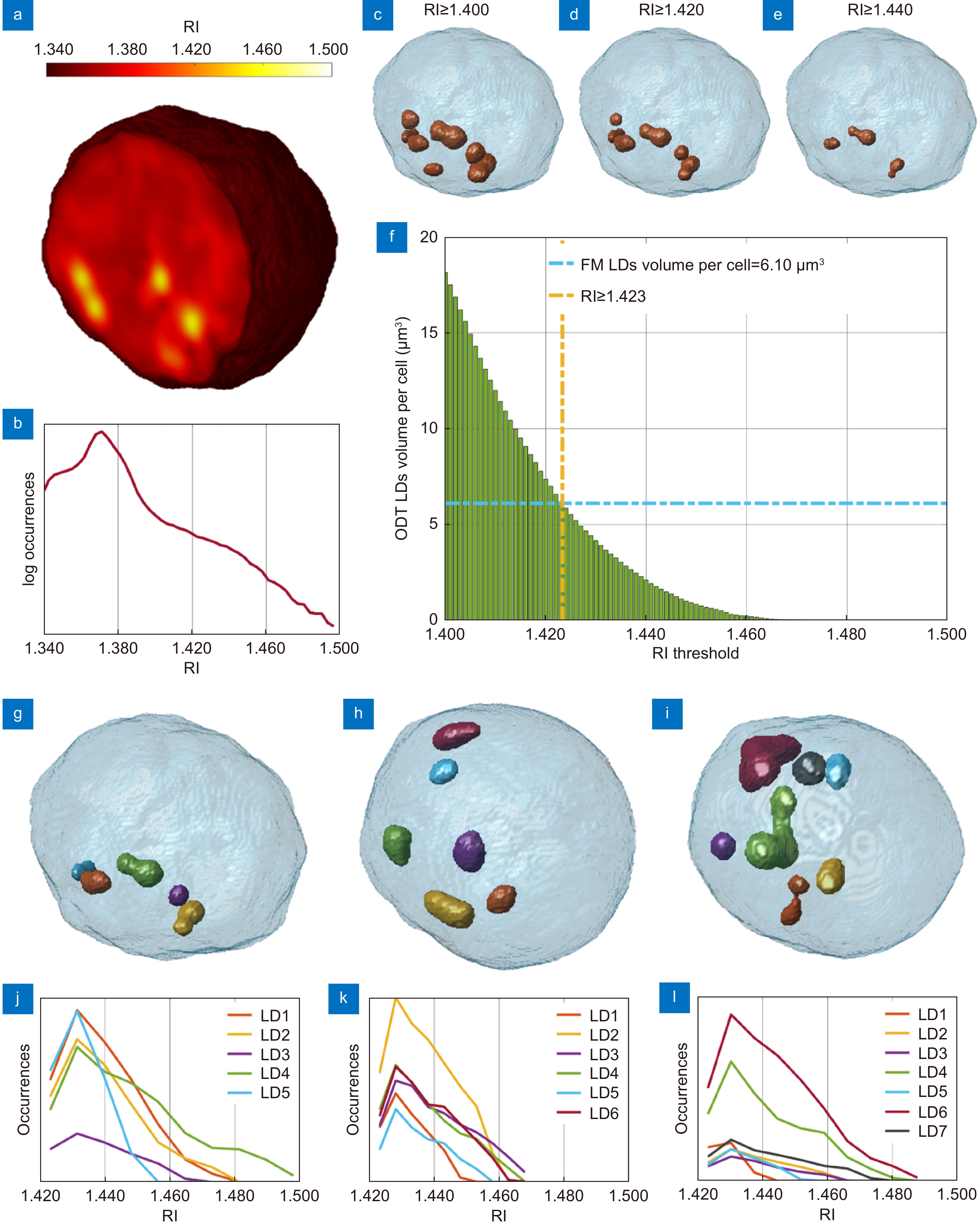
Figure 1.
Segmentation of the LDs within the 3D RI tomograms of A2780 live cells. (a) Central slice of the 3D RI tomogram of an A2780 live cell, in which LDs take the highest RI values. (b) Histogram in logarithmic scale of the 3D RI distribution of the cell in (a). (c–e) Isolevels representation of the tomogram in (a), in which LDs (orange) have been segmented by using the RI thresholds reported above. (f) Average volume per cell of the LDs segmented in 54 TPM tomograms of A2780 live cells by using different RI thresholds. The selected LDs-threshold (yellow line) allows computing the same average volume measured in 2D FM images (blue line). (g–i) Isolevels representation of separated LDs or LDs clusters segmented in 3 A2780 tomograms by using the LDs-threshold selected in (f), and (j–l) corresponding RI histograms. (a–e,g–j) are the same cell.
-
Figure 2.
LDs features extracted from 54 A2780 3D RI tomograms. (a–e) Histograms of respectively the mean value, standard deviation, entropy, kurtosis, and skewness of the 3D RI distributions of each LD. (f–h) Histograms of respectively the equivalent radius, the dry mass, and the sphericity of each LD. (i) Histogram of the distance between each LD centroid and the corresponding cell centroid, normalized to the cell equivalent radius. (j) Histogram of the distance between each LD centroid and the centroid of all the LDs inside the same corresponding cell, normalized to the cell equivalent diameter. (k) Mean RIs (orange dots) of concentric inner zones (orange regions) selected inside the same LD, with overlapped in blue the parabolic fitting. (l) Bivariate histogram of the first and second order coefficients of the parabolic fitting in (k) measured in all the LDs (black dots).
-
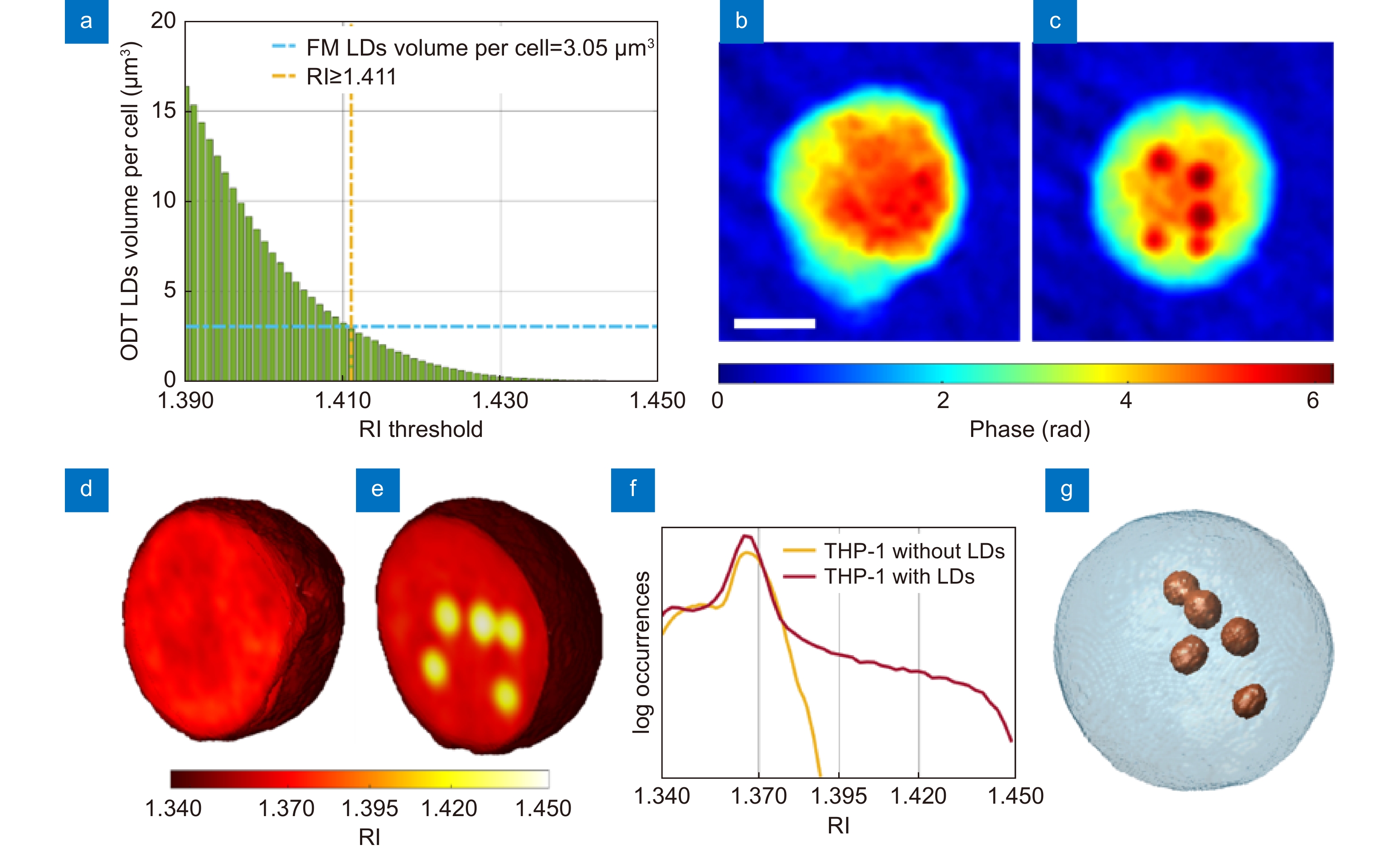
Figure 3.
Segmentation of the LDs within the 3D RI tomograms of THP-1 live cells. (a) Average volume per cell of the LDs segmented in 34 TPM tomograms of THP-1 live cells by using different RI thresholds. The selected LDs-threshold (yellow line) allows computing the same average volume measured in 2D FM images (blue line). (b, c) QPMs of two THP-1 cells, one without LDs (b) and the other one with LDs (c) (dark red spots). Scale bar is 5 μm. (d, e) Central slices of the 3D RI tomograms of the cells in (b,c), respectively, in which LDs take the highest RI values (Supplementary Movie 1). (f) Histogram in logarithmic scale of the 3D RI distribution of the cells in (d) (yellow) and (e) (red). (g) Isolevels representation of the tomogram in (e), in which LDs (orange) have been segmented by using the LDs-threshold selected in (a) (Supplementary Movie 2).
-
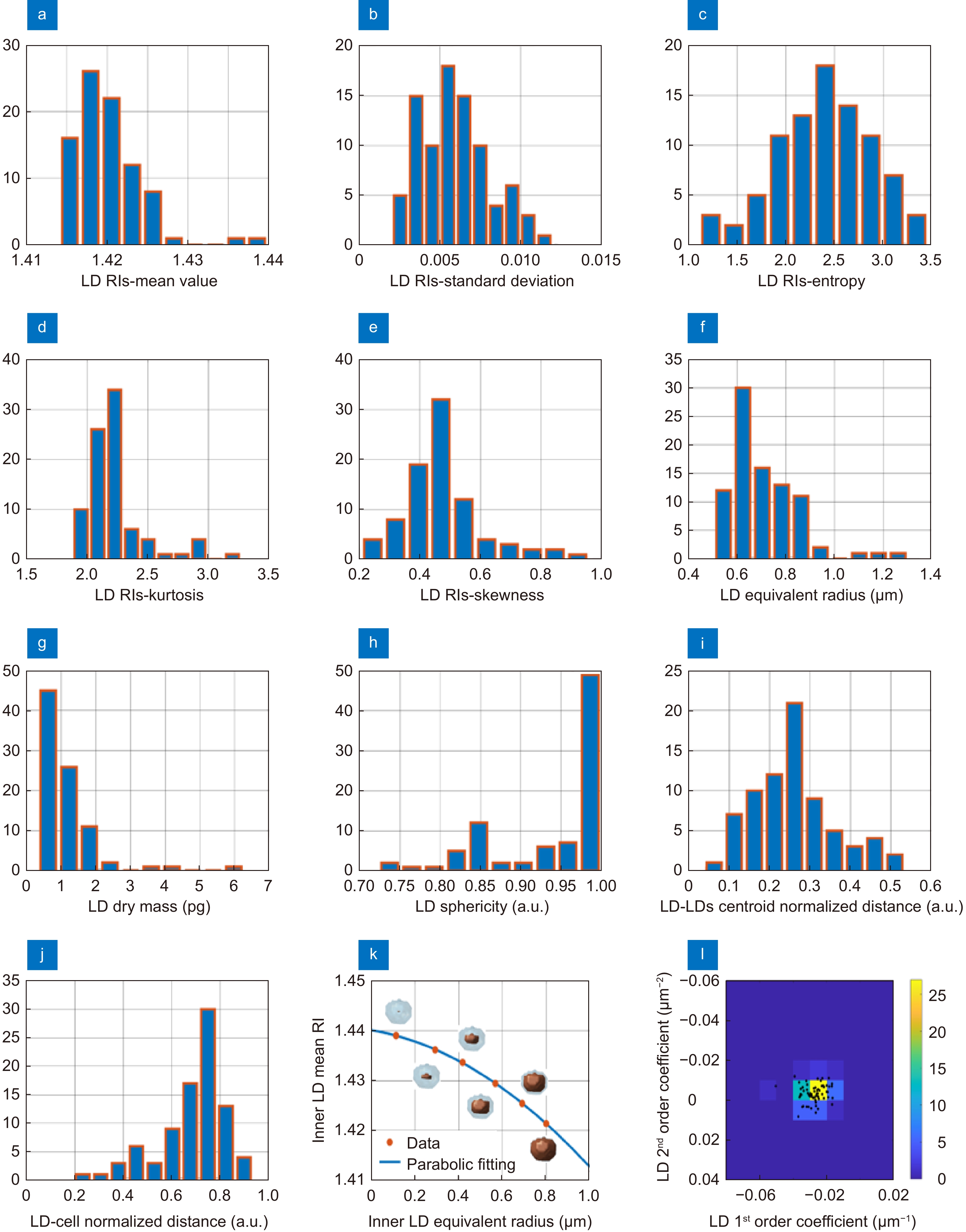
Figure 4.
LDs features extracted from 34 THP-1 3D RI tomograms. (a–e) Histograms of respectively the mean value, standard deviation, entropy, kurtosis, and skewness of the 3D RI distributions of each LD. (f–h) Histograms of respectively the equivalent radius, the dry mass, and the sphericity of each LD. (i) Histogram of the distance between each LD centroid and the corresponding cell centroid, normalized to the cell equivalent radius. (j) Histogram of the distance between each LD centroid and the centroid of all the LDs inside the same corresponding cell, normalized to the cell equivalent diameter. (k) Mean RIs (orange dots) of concentric inner zones (orange regions) selected inside the same LD, with overlapped in blue the parabolic fitting. (l) Bivariate histogram of the first and second order coefficients of the parabolic fitting in (k) measured in all the LDs (black dots).
-
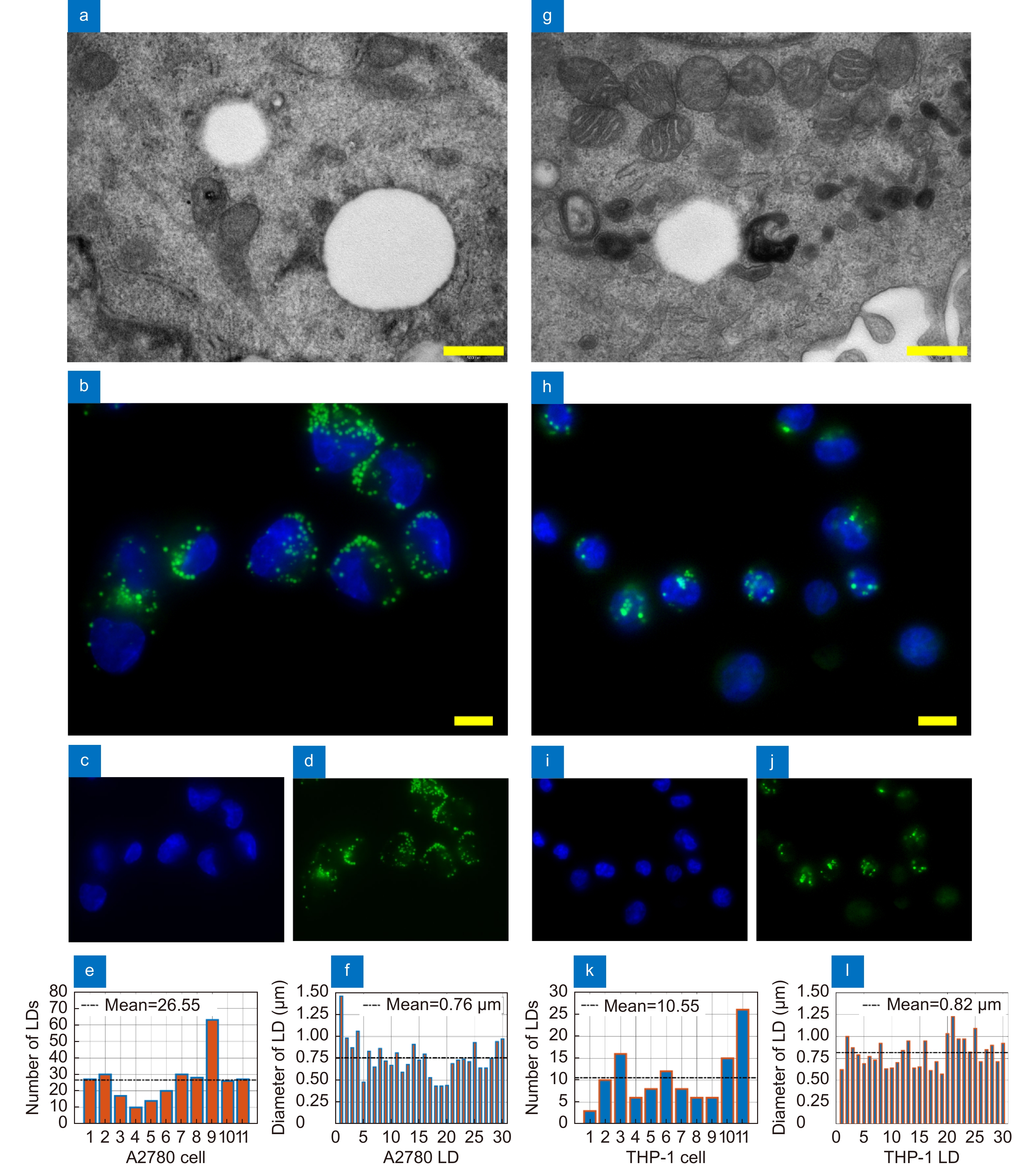
Figure 5.
Conventional 2D imaging of A2780 (a–f) and THP-1 (g–l) live cells. (a, g) TEM images of LDs. Scale bar is 0.5 μm. (b, h) Representative FM images of nuclei (blue) and LDs (green). Scale bar is 10 μm. (c, i) FM images of nuclei stained with Hoechst. (d, j) FM images of LDs stained with Nile Red. (e, k) Number of LDs in 11 live cells imaged by FM. (f, l) Diameters of 30 LDs imaged by FM.
-
Figure 6.
In-flow TPM system. (a) DH microscope in off-axis configuration. PBS – polarizing beam splitter; WP –wave plate; M – mirror; L1, L2 – Lens; MO – microscope objective; MC – microfluidic channel; MP – microfluidic pump; TL – tube lens; BS – beam splitter; CMOS – camera. (b–d) Three QPMs of an A2780 cell while flowing along the y-axis and rotating around the x-axis. The spots with the biggest phase values (dark red) are the LDs. Scale bar is 5 μm. (e–g) Pseudo-3D visualization of the 2D QPMs in (b–d), respectively, in which the LDs are well-separated from the outer cell because of their greater height.
-
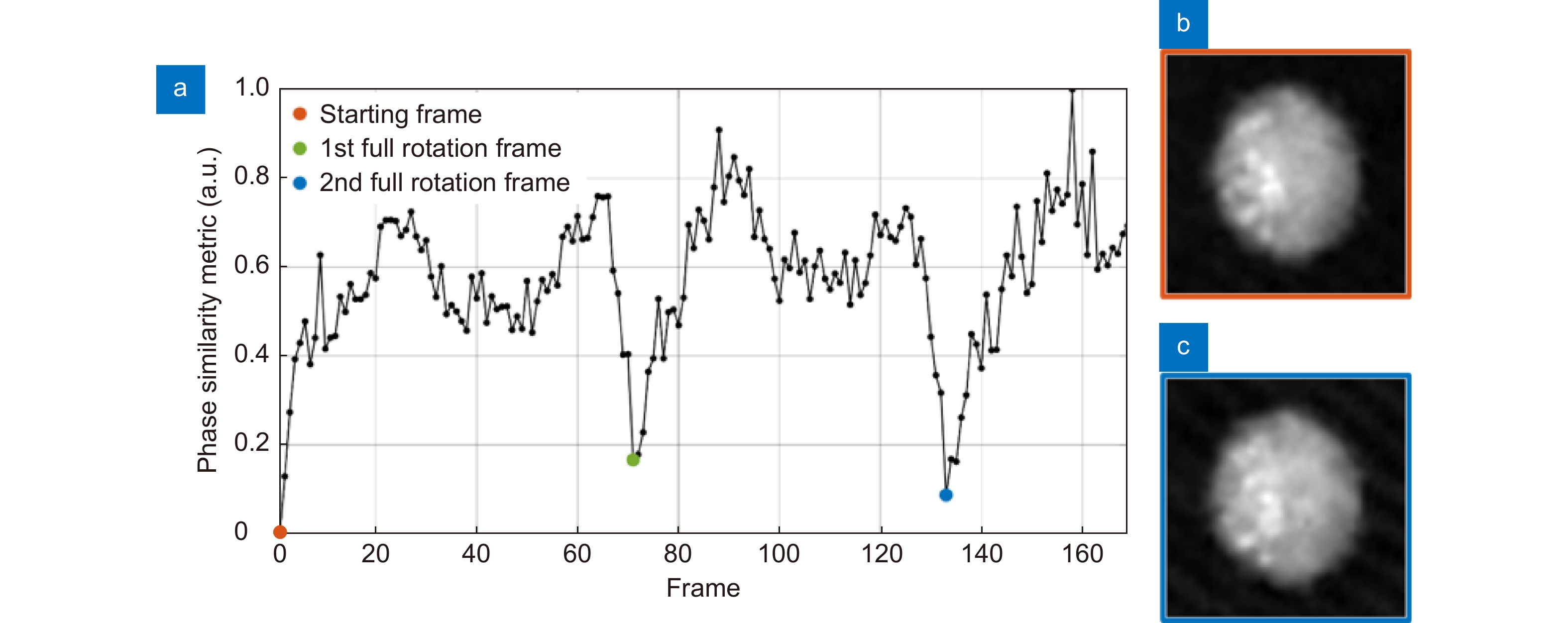
Figure 7.
LDs-aided method for the rolling angles recovery in an A2780 live cell. (a) Trend of the phase similarity metric, which is null in the starting frame of the QPM sequence (orange dot) and is minimum when the first (green dot) and the second (blue dot) full cell rotations have occurred. (b) QPM at the first frame and (c) QPM after two full rotations, in which LDs are located in the same positions.

 E-mail Alert
E-mail Alert RSS
RSS

 DownLoad:
DownLoad: